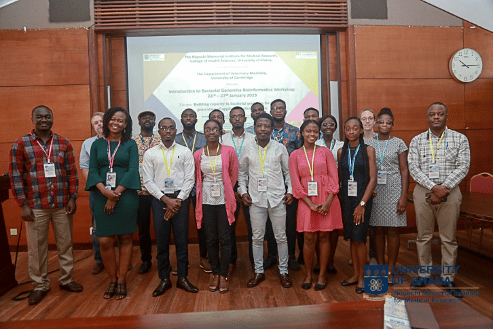
A five-day intensive training on Introduction to Bacterial Genomics Bioinformatics held from 23rd to 27th January 2023 has been organised at the Noguchi Advanced Research Laboratory (ARL) bioinformatics training room. This is the first in its series resulting from a collaboration between the Noguchi Memorial Institute for Medical Research, College of Health Sciences – University of Ghana (NMIMR-UG), and the Department of Veterinary Medicine – University of Cambridge, with support from the Africa Research Excellence Fund (AREF) and the Cambridge – Africa ALBORADA Research Fund.
Instructors and facilitators for this maiden workshop included Dr. Andries van Tonder (instructor) from the University of Cambridge, Dr. Prince Asare (instructor), Dr. Bright Adu (facilitator), Dr. Isaac Darko Otchere (facilitator), Dr. Benedicta Ayiedu Mensah (facilitator) and Mr. Kwesi Agyenkwa-Mawuli (facilitator), all from NMIMR.
This workshop began with a short opening ceremony during which the Director of the Institute, Professor Dorothy Yeboah-Manu, welcomed all facilitators and invited guests and wished them a successful workshop. She expressed the hope that by increasing our knowledge, we may help the Institution and trainees grow. The participants of the workshop were happy to receive two key seminars on bioinformatics given by experts in the field – on the morning of day one by Professor Julian Parkhill, from the Department of Veterinary Medicine – University of Cambridge and the morning of day four by Dr. Marco van Zwetselaar from Kilimanjaro Clinical Research Institute, Tanzania.
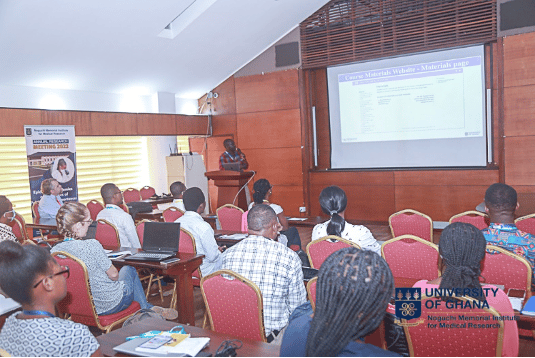
The workshop-built capacity in bacterial genomics for the next generation of applied bioinformaticians. The training module also concentrated on the differences between sequencing data produced by Illumina and Nanopore platforms, recognizing the structure of common file formats in bioinformatics particularly in FASTA and FASTQ files and introduced participants to the use of FastQC to MultiQC to produce quality reports for Illumina sequences.

There were hands-on sessions on genome assembly and annotation, workflow managers and pipelines, and Drs. Asare and van Tonder also led participants through introduction to Unix and the command line, common bioinformatics file formats, sequencing QC, short read mapping, introduction to phylogenetics as well as anti-microbial resistance prediction form sequencing reads.

In all, there were 15 participants who benefited from the training that was led by the various Instructors and facilitators. Participants learning were enhanced, and all expressed great gratitude for being part of the training.